Debunking myths on genetics and DNA
Musings on writing, genetics and photography. Sign up for my newsletter to get notified of promotions, ARCs, and free stories.
Saturday, December 17, 2016
We are the "educated elite" they won't listen to
Why did Trump win the 2016 presidential election? Since November 9, many people have been asking the same question. Some say it was because Clinton wasn’t likable enough. Others blame the fact that we didn’t understand the white working class and we believed too many fake news and conspiracy theories.
Yes, those are just some of the answers. There are many more, of course, and political scientists and historians will debate this election for decades to come. But I’ve reached a point where I’ve had enough of reading how everyone else is feeling about this election. I need to say how I feel about it. Because I’m not the white working class woman who lost her job, I’m not Muslim, I’m not Mexican, and yes, I’m an immigrant but the privileged kind if you will. I’ve always been legal and now I am, in fact, a citizen.
I am the educated elite to whom it’s ok to say, “Your college degree doesn’t make you smarter.” The one who tries to counter-argue using scientific sources, real numbers, and logic, and yet all she gets back in response is name-calling and ridicule.
And if you are reading this blog, I know you are part of that same group of people who strive to educate themselves and improve their knowledge, the group of people now clumped under the umbrella term of "educated elite." So you know exactly what it feels like to be in a strange society where beliefs overcome centuries of scientific and ideological progress. It feels like the doctor who’s been telling his patients to go on a diet and quit eating junk food. We all want to be healthy and fit, yet when it boils down to making the effort, many shrug off the scientific evidence that bad eating habits and no exercise harm our health. We’re all gonna die anyway, right?
No, a college degree doesn’t make anyone smarter. Critical thinking does. And while for many things a college degree is not even needed, there are others—like health, science, and the environment for example—for which those extra years of education provide perspective and deeper understanding of those fields in particular. So when a person with a college degree or equivalent experience tells us something about the field they've studied, I think we should listen. What kind of society have we become when we no longer trust scientists when they tell us that greenhouse gases are causing the ocean temperatures to rise? Or when we no longer listen to our doctors when they say that a vaccine can save our children’s life?
There was a time when education was revered and school teachers were respected. A time when people listened to educators and doctors because they had spent decades studying and gaining their knowledge. A time when scientists were heroes because they got us to the moon and physicians saved lives with vaccines. Today, I tell my kids to study because otherwise they’ll never get a good job, and what do they reply? “Mom, Mark Zuckerberg and Bill Gates dropped out of college.” Or, even better: “Mom, YouTubers make millions of dollars and they never have to go to college.”
The Internet has it all. Because anyone can contribute to the Internet, it has all the answers your doctor will never give you, all the science your religion allows you to believe, and a perfect world that beautifully matches your so called values. Why bother with education? Evolution isn’t real, it’s just a theory. We don’t need educated people telling us how old the earth is. We don’t need educated people injecting stuff in our kids’ arms claiming it’ll save their lives. We don’t need educated people telling us that the climate is changing—it’s been changing all throughout the 6,000 years the earth has been around.
Just so we’re clear, what I did in those last three sentences is called sarcasm. Because I did go to school, for many years, in fact. I went to college and then to graduate school. I got my PhD while I was raising two young children. I worked my ass off, I suffered through many failures, I received many rejection letters and yet I kept plowing along, learning from my mistakes, working through the adversities. No, you don't need a college degree to achieve that. Any kind of hard work makes people stronger. Learning from our mistakes and taking responsibilities makes us grow. And it teaches to respect one other.
But no, America doesn’t need any of that. America keeps binging on junk food and relying on whatever the Internet has to offer. So now teachers are indoctrinating our kids, scientists are conspiracy theorists who enjoy telling us that the world will end, and doctors are vaccine impostors paid by the Big Pharma. (On a side note, vaccines are cheap, they don’t make the Big Pharma rich. What makes them rich are all the drugs you need when you get the diseases you could’ve vaccinated against.)
Who needs educated people when we can do stuff on our own? This, you see, has been this election’s winning message. And that’s exactly why the incoming cabinet features a secretary of education who wants to dismantle public education; a national security adviser who tweets fake news; a secretary of treasury who ran a bank that was dubbed the “foreclosure machine”; an EPA administrator who doesn’t believe in environmental policies and doesn’t believe in climate change. If you think about it, it’s like all these people, instead of being rightfully shamed for their failures, have been rewarded with the highest positions in the government. All because we no longer trust education, let alone if it comes from the establishment. Can you hear your teenage kid’s voice? “Get out of the way, Mom. I can do stuff on my own, now!”
Truth is, the secretary of energy for the past two terms has been a PhD. In fact, our previous secretary of energy was a Nobel laureate. These bright minds will now be succeeded by a BS in animal science and Dancing with the Stars contestant. I’m sure those stardom experiences will come in handy when talking to scientists and engineers about the state of our nuclear weapons and climate change policies. Oh wait. I forgot, the guy doesn’t believe in climate change. So I’m sure he doesn’t believe in investing money in basic research to address questions like, “Why are our trees dying?” or “Is there going to be enough water for our growing cities twenty years from now?” or “How are we going to sustain our agriculture when we’ll run out of water?” or “Can we make more potent antibiotics in order to fight drug resistant bacteria?”
And if you are wondering, all those questions do pertain the US Department of Energy. DOE oversees national security labs like the one where I work. My colleagues and I work hard everyday on questions like the ones I mentioned above because drug-resistant bacteria and water problems are among the greatest threats our society is facing today. Ill-minded people can easily get a hold of these problems and turn them into bioweapons. But don’t worry, I’m sure the Internet holds all the answers.
Nelson Mandela once said, "Education is the most powerful weapon which you can use to change the world." I want you to think about that for a moment. What do Mandela's words say about a country where college education is so expensive that many can no longer afford it? And even worse, a country where a good chunk of the population does not trust college education? Knowledge is freedom. Ignorance, instead, is the shackles used by tyrannic powers.
And so America gulps down another mouthful of fat hamburger with fries, washed down with corn syrup based soda. Because no matter how many times we tell America to go on a diet (i.e. invest in renewable energy), no matter how many times we lecture America about the risks of high blood pressure (i.e. stop fracking and drilling before all of our drinking water ends up contaminated), and no matter how many times we remind America that the number one cause of death in the US is heart failure (i.e. temperatures are indeed rising and we’re all going to die if we don’t do something about it), America has decided that we are the educated elite and no longer deserve to be listened to.
So good luck America. In a world too busy to take the time to ponder, too loud to stop to listen, and too arrogant to realize its own ignorance, the educated elite is all you’ll have left when catastrophe will strike. And in the best of Hollywood traditions, we will be there to come to the rescue.
Until then, God help us all.
Thank you to Kat Fieler and Mike Martin for helpful suggestions while editing this post. All opinions expressed here are mine and mine alone. I respect yours, please respect mine.
Labels:
random musings
Wednesday, October 5, 2016
Hello All, happy fall and happy Halloween month!
First off, if you haven't heard about the anthology contest, check it out: the deadline is November 1st! Rules, theme and other important details on the ISWG page.
My post this month is going to be short and I apologize if I won't be able to reciprocate the comments until later in the week since I'm currently at a conference (and my presentation is today, wish me luck!).
October question: When do you know your story is ready?
It's hard to give an objective answer to that question since a story is ready when it "feels" ready. But since for a writer the hardest thing is to judge his/her own work, my strategy has been to write a first draft, then go back and refine, then go back and edit, then go back and send it to trusted beta readers. Sometimes a beta reader will come up with a suggestion that does not resonate, but most of the times, my trusted betas have good suggestions and after those improvements I usually feel that the story is ready.
How about you, what's your strategy?
Labels:
IWSG
Friday, September 9, 2016
Pictures from the California Coast
I love the Pacific coast. It's simply gorgeous from Mexico all the way to Alaska. Over my last trip to California I was rewarded with some really nice sunsets. Here are my latest pictures, and as always, you can get prints from my website www.elenaegiorgi.com
Enjoy!
And the best part? As we were strolling along the beach in Carmel we spotted bottlenose dolphins! Aren't they adorable??
Enjoy!
 |
| Goleta, CA |
 |
| Goleta, CA |
 |
| Cambria, CA |
 |
| Cambria, CA |
 |
| Pebble Beach, CA |
 |
| Point Lobos, CA |
And the best part? As we were strolling along the beach in Carmel we spotted bottlenose dolphins! Aren't they adorable??
Labels:
My photos
Wednesday, September 7, 2016
September IWSG: have you been taking care of your newsletter?
Hello fellow writers, my big news this month is that we launched the new anthology Beyond the Stars: at the Galaxy's Edge, featuring my story The Quarium Wars, and it was a great success! I'm really excited and stoked to be part of this project. Now onward to finish the book that's actually set in the world of my short story.
Which brings me to this month's question: How do you find the time to write in your busy day?
My answer is very simple. I don't. Fall is particularly busy for me because on top of my daily work I get a lot of requests for portrait session -- which is VERY good, don't get me wrong, I LOVE doing portrait sessions -- and so guess what happens to my manuscripts? They're left behind. Le sigh.
In other news, I'm working on revamping my website and newsletter. Newsletters are very important when you are trying to grow your business. In fact, they are like plants: you have to keep watering and nurturing it. There are many providers out there, and so far I've used MailChimp, which has been very good except for two drawbacks: (1) emails to gmail users go to the promotional tab and may never be read/seen; (2) when you start getting more than 1,000 subscribers it becomes very expensive.
So I've looked into other providers, but it's a bit of a pain migrating. The good news is that for most of them you can sign up for free and test them out before you start paying. What provider do you use? And are you happy with it?
Until next time!
Labels:
IWSG
Wednesday, August 3, 2016
August IWSG post
Howdy, I'm back after some traveling, which is why I was absent last month (apologies). I hope everyone in the group is having a wonderful summer (winter for those of you in the southern hemisphere). And if the heat and nice weather is keeping you from writing (*coughs*), be nice to yourself because you never know where your next inspiration will be. Maybe your muse is calling from the beach, what do you know? ;-)
Jokes aside, I've been beating myself up, actually, because all I've produced this year so far are three short stories, and while two of them were slated to go in anthologies, both anthologies have been delayed. So, if you are in a similar situation, don't despair: here's my good news that may inspire you and keep you from giving up. A wonderful opportunity opened up last month and my third story was accepted for an anthology that's coming out this month, so yay! And one of those two anthologies that got delayed was mentioned recently on the TOR website as one of the 15 books to watch for in 2016 -- so double YAY! (It's at the very bottom, Zero Machine by Acheron Books, but still, the fact that it was even mentioned is super cool, I was happy dancing all over the place!)
So now I've to go back to that space opera that I was supposed to finish two months ago... What about you? What projects are keeping you busy these days? Stay inspired!
On a different note: Together with a bunch f authors we are hosting a mega-giveaway with lots of prizes, including a $50 Amazon gift card! For more details please see this post.
Friday, July 29, 2016
The Abandoned Asylum
I had the unique privilege to visit an abandoned asylum and here are the images I made so far. I have so many more, I will be busy compositing until next year. :D
For the above I downloaded the wings made by Deviant Art artist "Thy Darkest Hour".
For the above I downloaded the wings made by Deviant Art artist "Thy Darkest Hour".
Labels:
My photos
Sunday, July 24, 2016
Sci-Fi Summer Fling, win a $50 gift card and lots of sci-fi books!
Who doesn't love a good Summer Fling? Entire Musicals have been written about it! And today we're here to bring you the Summer Fling of a Book Lover's dreams. Twenty SciFi writers have come together to offer a multi-media book experience for bibliophiles that you just can't resist. These authors include Hugo Nominees, International Best-Sellers, Award Winners, USA Today Best-Sellers, and some of the Best Sci-Fi Authors available today (even NPR agrees!)
We have paperbacks, ebooks, audiobooks, swag, a $50 gift card and more, all for ONE lucky winner. Sounds amazing, doesn't it? The event is from 7/23 and 8/6 so don’t miss the chance to enter!
Check out the giveaway here: http://www.pktyler.com/SFling
Enter and you have a chance to win gifts from these authors, plus you'll be added to their newsletters where you'll have more chances to get free ebooks and hear about new releases and other promotions.
Prizes include:
$50 Amazon Gift Card
Paperbacks of: Scavenger: Evolution, Beyond the Stars: A Planet Too Far, Massacre at Lonesome Ridge, The Luna Deception, The Future Chronicles - Special Edition, Shroud of Eden, Paperback of Strikers, First 2 paperbacks in the Corporate Marines series, The Running of the Tyrannosaurs, Blink, Noah Zarc: Mammoth Trouble, Rebel's Honor
Audible copy of: Red Rex: Blood Echoes
Swag: KN Lee Tote bag
Ebooks of: e-book of Paradisi Escape, Book 1 of the Paradisi Exodus Series, ebook copy of The Morph (Gate Shifter Book 1), ebook of The Invariable Man
So don't hesitate, enter now (http://www.pktyler.com/SFling) and keep sharing for more chances to win!
Thank you from the Sponsoring Authors:
Gwynn White author of Rebel's Honor
Demelza Carlton with a new release coming this fall
D. Robert Pease author of Noah Zarc: Mammoth Trouble
Timothy C. Ward author of Scavenger: Evolution
Patrice Fitzgerald Publisher of Beyond the Stars: A Planet Too Far
Samantha Warren author of Space Grease & Pixie Dust
Felix R. Savage author of The Luna Deception
Cheri Lasota author of Paradisi Escape
Samuel Peralta publisher of The Future Chronicles - special edition
P.K. Tyler author of The Jakkattu Vector
Marlin Desault author of Shroud of Eden
Ann Christy author of Strikers
Tom Germann author of Corporate Marines
Calinda B author of Red Rex: Blood Echoes
Stant Litore author of The Running of the Tyrannosaurs
JC Andrijeski author of Rook (Allie's War Book One)
E. E. Giorgi author of Akaela (Dystopian YA)
A.K. Meek author of The Invariable Man
Will Swardstrom author of Blink
K.N. Lee author of Spell Slinger
Friday, June 3, 2016
I'm in love with our mountains
Another post on the beauty of the Jemez Mountains, in Northern New Mexico. For the record, I actually grew up by the sea and I miss my morning strolls along the beach. But it's hard not to fall in love with these views. This was by the San Antonio River. The water flows at the bottom of a canyon, and the cliffs around them yield stunning views.
I didn't upload all of the pictures on my website, so if you are interested in prints please email me (contact info here).
As the sun went down it started drizzling and the fine layer of clouds diffused the sunset light creating beautiful pink hues in the sky. It was magical! :-)
I didn't upload all of the pictures on my website, so if you are interested in prints please email me (contact info here).
 |
| © Elena E. Giorgi |
 |
| © Elena E. Giorgi |
As the sun went down it started drizzling and the fine layer of clouds diffused the sunset light creating beautiful pink hues in the sky. It was magical! :-)
 |
| © Elena E. Giorgi |
 |
| © Elena E. Giorgi |
Labels:
My photos
Wednesday, June 1, 2016
June IWSG: an inspirational story
This month I want to share a little episode that happened to me because I found it to be a great inspiration, and I hope you'll find it inspirational too.
You have to know that I'm not exactly pretty. Never been. Let's just say that beauty is not my gift. But, but, but, I'm healthy, and I have no missing body parts. I would never replace or cut or mar in any way my appearance in the name of beauty because there are so many people out there who are missing a leg or a hand or have crippling genetic conditions, so imagine how disrespectful it would be toward those people to go under the knife in the name of esthetics. So I compensate with art. I do photography, I write. I try to do beautiful things.
Wait, wait, I actually have a story to tell you, so don't start saying, "Awww, but you're beautiful inside," because you know what reply that will prompt: "Who the hell's gonna come and turn me inside out??" *grin*
Back to my story. I have a friend at work who's really beautiful. She has one of those perfect faces that never age and know no flaws. A few weeks ago we went out to lunch together and out of the blue she told me, "I envy you."
I almost fell off my chair. I said, "What are you talking about, Kate? Did you take a good look at me? And did you take a good look at the mirror? How can somebody as beautiful as yourself envy _me_?"
She shook her head. "You don't understand. This" -- she pointed to herself -- "I had nothing to do with this. I have only my parents to thank for their good genes. But you -- you have talents. And you take full credit for those talents."
I was taken by such surprise that I didn't know what to reply. So I hugged her and thanked her.
Why did I tell you this? Because we all hate some things of ourselves. We all have insecurities. But maybe sometimes we have to learn to look at ourselves with somebody else's eyes and be more forgiving. We are fairly good at forgiving others, so let's learn to do the same with ourselves. :-)
Labels:
IWSG
Friday, May 27, 2016
The viruses inside us
 |
| Dendogram of endogenous retroviruses. Source: Wikipedia. |
Last month I posted a discussion on a PNAS paper that reported the discovery of a new class of viruses, called pithoviruses, found in a layer of Siberian permafrost. In their paper [1], the researchers conclude:
"Our results further substantiate the possibility that infectious viral pathogens might be released from ancient permafrost layers exposed by thawing, mining, or drilling."I found this possibility intriguing both from a scientific point of view as well as a sci-fi point of view: there are plenty of books out there on zombies and aliens, but what about ancient viruses that thawed from the ice thanks to global warming?
An attentive reader, though, didn't buy the sci-fi "threat" and asked in the comments whether viruses are necessarily bad. Normally we think of viruses as pesky little things. And while most will make us sick for a short time only, some can indeed be deadly, and others can inflict long-term complications.
The reader who asked that question, however, is absolutely right: over the course of evolution, viruses have been beneficial to us. Viruses have driven genetic diversity by transferring genes across species, and in fact, we still carry remnants of viral genes in our DNA, comprising roughly 8-10% of our genome. They are called "endogenous retroviruses", or ERV.
In the rest of this post I will address two questions:
- What are those viral genes doing in our genome?
- How did they get there?
What are viral genes doing in our genome?
Most of them are doing nothing. They are "deactivated", meaning they do not code for proteins. Our genome is made of many redundant elements that over the course of evolution were silenced because no longer useful, only to be turned on again later on when a new adaptation happened.
One such example is the placenta, where endogenous retroviruses have been found to be expressed [2-4] and play a role in the growth and implantation of the tissue. We can only speculate on why retroviral genes are expressed in the placenta, but the hypothesis is indeed quite interesting: in order to survive, retroviruses debilitate the immune system. In general, this is not a good thing for the body, except in one very special instance: an embryo is literally a parasite growing inside the mother's body. It carries extraneous DNA and, under normal circumstances, something carrying extraneous DNA would be considered an antigen and attacked by the immune system. Therefore, the expressed viral proteins found in the trophoblasts, the outer layer of the placenta, would have the role of suppressing a possible immune reaction against fetal blood.
Another property viruses have is that of cell fusion: they literally "merge" cells together into one membrane. A second hypothesis is that this property is used during the development of the placenta to build a barrier between the maternal circulation and the fetal circulation.
How did viral genes end up in our genome?
A virus enters the body of a host with the sole purpose of replicating. In order to do so, viruses hijack the cell's own replicating machinery. Retroviruses in particular carry strands of RNA which, once injected inside the cell, are turned into DNA that is then carried inside the cell nucleus and integrated into the cell's genome. This ensures that once the cell replicates, the bit of viral DNA is replicated too.
There is a special set of cells, however, such that when the virus infects them it literally gets stuck. These cells are the gametocytes, a.k.a. oocytes in women, and spermatocytes in men, which do not duplicate unless they get fertilized. But by then the virus is no longer active. It's literally stuck, in the sense that the integrated viral DNA now cannot replicate and cannot escape the host's DNA. It's just a bit of non-functional DNA that gets duplicated along as the embryo grows. The new individual now carries the viral genes in every cell of his/her body, even in the gametocytes, and hence the viral genes will be inherited by future generations as well.
And that's how viruses ended up in our genome a long, long time ago and have literally become "evolutionary fossils." In fact, by looking at these endogenous retroviral sequences, scientists are able to reconstruct the evolution of ancient viruses.
References
[1] Legendre, M., Bartoli, J., Shmakova, L., Jeudy, S., Labadie, K., Adrait, A., Lescot, M., Poirot, O., Bertaux, L., Bruley, C., Coute, Y., Rivkina, E., Abergel, C., & Claverie, J. (2014). Thirty-thousand-year-old distant relative of giant icosahedral DNA viruses with a pandoravirus morphology Proceedings of the National Academy of Sciences, 111 (11), 4274-4279 DOI: 10.1073/pnas.1320670111
[2] Emerman M, & Malik HS (2010). Paleovirology--modern consequences of ancient viruses. PLoS biology, 8 (2) PMID: 20161719
[3] Dunlap KA, Palmarini M, Varela M, Burghardt RC, Hayashi K, Farmer JL, & Spencer TE (2006). Endogenous retroviruses regulate periimplantation placental growth and differentiation. Proceedings of the National Academy of Sciences of the United States of America, 103 (39), 14390-5 PMID: 16980413
[4] Dupressoir A, & Heidmann T (2011). [Syncytins - retroviral envelope genes captured for the benefit of placental development]. Medecine sciences : M/S, 27 (2), 163-9 PMID: 21382324
Labels:
DNA,
Evolution,
pseudogenes,
virus
Sunday, May 15, 2016
To all of you who think New Mexico is all desert ...
... think again! ;-)
The Jemez mountains are covered in ponderosa pines and rivers like Rio Puerco give rise to the beautiful waterfalls we scouted with some photographer friends this past week-end. Enjoy!
The Jemez mountains are covered in ponderosa pines and rivers like Rio Puerco give rise to the beautiful waterfalls we scouted with some photographer friends this past week-end. Enjoy!
 |
| © Elena E. Giorgi |
 |
| © Elena E. Giorgi |
 |
| © Elena E. Giorgi |
 |
| © Elena E. Giorgi |
Labels:
My photos,
New Mexico
Friday, May 13, 2016
Using Supercomputers to Probe the Early Universe
 |
| Artist's depiction of the WMAP satellite gathering data to understand the Big Bang. Source: NASA. |
For decades physicists have been trying to decipher the first moments after the Big Bang. Using very large telescopes, for example, scientists scan the skies and look at how fast galaxies move. Satellites study the relic radiation left from the Big Bang, called the cosmic microwave background radiation. And finally, particle colliders, like the Large Hadron Collider at CERN, allow researchers to smash protons together and analyze the debris left behind by such collisions.
Physicists at Los Alamos National Laboratory, however, are taking a different approach: they are using computers. In collaboration with colleagues at University of California San Diego, the Los Alamos researchers developed a computer code, called BURST, that can simulate a slice in the life of our young cosmos.
While BURST is not the first computer code to simulate conditions during the first few minutes of cosmological evolution, it can achieve better precision by a few orders of magnitude compared to its predecessors. Furthermore, it will be the only simulation code able to match the precision of the data from the Extremely Large Telescopes currently under construction in Chile. These new telescopes will have primary mirrors that range in aperture from 20 to 40 meters, roughly three times wider than the current very large telescopes, and an overall light-collecting area up to 10 times larger.
A few seconds after the Big Bang, the universe was composed of a thick, 10-billion degree "cosmic soup" of subatomic particles. As the hot universe expanded, these particles' mutual interactions caused the universe to behave as a cooling thermonuclear reactor. This reactor produced light nuclei, such as deuterium, helium, and lithium — all found in the universe today. "Our code, developed with Evan Grohs, who at the time was a graduate student at UCSD, looks at what happened when the universe was about 1/100 of a second old to a few minutes old," says Los Alamos physicist Mark Paris of the Theoretical Division. "By determining the amount of helium, lithium and deuterium at the end of those first few minutes of life, BURST will be able to shed light to some of the existing puzzles of cosmology."
One such puzzle is dark matter: physicists know that such matter exists because of the way galaxies rotate, but they haven't been able to detect it because it does not radiate in any known spectrum. Physicists have theorized that dark matter is made of so-called "sterile neutrinos", which do not interact with any other particle and are responsible for these unobservable interactions. "Once we start getting data from the Extremely Large Telescopes," Paris explains, "we will model sterile neutrinos into the BURST code. If we get a good description, we will be able to prove their existence."
Measurements of the cosmic microwave background radiation have led physicists to theorize "dark radiation," a speculative form of energy that may have acted in the early universe. BURST could possibly reveal whether or not dark radiation is real and caused by sterile neutrinos. "The universe is our laboratory," Paris enthusiastically concludes. "BURST will help us answer questions that are currently very difficult to address with particle colliders like the one at CERN."
Ongoing support for the project is provided by the National Science Foundation at UCSD and the Laboratory Directed Research and Development program through the Center for Space and Earth Sciences at Los Alamos. BURST will be running on the supercomputing platforms at Los Alamos National Laboratory.
Reference
Grohs, E., Fuller, G., Kishimoto, C., Paris, M., & Vlasenko, A. (2016). Neutrino energy transport in weak decoupling and big bang nucleosynthesis Physical Review D, 93 (8) DOI: 10.1103/PhysRevD.93.083522
Wednesday, May 4, 2016
May IWSG: Spring is finally here!
First off a big announcement: the IWSG anthology, titled Parallels: Felix Was Here, is here! Featuring 10 stories from ISWG authors, hand-picked by a panel of agents and writers, you can now get it from Amazon and other retailers. Complete list of purchasing links here.
It's May already, can you believe it? How was your month of April, did you do the A-Z challenge? Already making plans for the summer?
My April wasn't too bad: I'm wrapping up a project at work and I finished a short story which will be the prequel to a new world I'm creating. It's a space opera and I'm very excited about it mostly because... I've never written space opera before! :-) Here's a question for you: would you release a prequel as soon as it's ready or would you wait until the first installment in the series is ready to be released too?
In other news, we've had some very much needed moisture in Northern New Mexico and so I took the chance and shot some droplet macros. :-) Happy May everyone!
Friday, April 29, 2016
Hunting For The Signatures of Cancer
 |
| Signatures of Mutational Processes Extracted from the Mutational Catalogs of 21 Breast Cancer Genomes. Credit:http://dx.doi.org/10.1016/j.celrep.2012.12.008 |
Cancer is the second leading cause of death worldwide, with approximately 14 million new cases and 8.2 million cancer related deaths each year (Source: WHO). A family history of cancer typically increases a person's risk of developing the disease, yet most cancer cases have no family history at all. This suggests that a combination of both genetics and environmental exposures contribute to the etiology of cancer. In this context, "genetics" means the genetic make-up we are born with and inherited from our parents. For example, women born with specific mutations in the BRCA1 and BRCA2 genes are known to have a much higher risk of developing breast cancer later in life.
However, besides the genetic make-up we carry from birth, there are many geographical and environmental factors that contribute to the risk of cancer. For example, the incidence of breast cancer is over 4 times higher in North and West Europe compared to Asia and Africa (Source: WHO). Stomach cancer, on the other hand, is much more prevalent in Asia than the US. If you think that this may be linked to the genetic differences across ethnicities, think again. The National Cancer Institute published a summary of several studies that compared the incidence of first and second generation immigrants in the US with the local population. They found that:
"cancer incidence patterns among first-generation immigrants were nearly identical to those of their native country, but through subsequent generations, these patterns evolved to resemble those found in the United States. This was true especially for cancers related to hormones, such as breast, prostate, and ovarian cancer and neoplasms of the uterine corpus and cancers attributable to westernized diets, such as colorectal malignancies."According to the World Health Organization (WHO),
"around one third of cancer deaths are due to the 5 leading behavioral and dietary risks: high body mass index, low fruit and vegetable intake, lack of physical activity, tobacco use, alcohol use."Cancer is the result of a series of cellular mechanisms gone awry: every time a cell divides, somatic mutations accumulate in the cell's genome. These are not mutations we are born with, inherited from our parents. Rather, these are changes that accumulate in certain cells as we grow old and are not the same across all cells in the body. Many environmental exposures contribute to this process and affect the rate at which these mutations accumulate. However, cells have various mechanisms that are normally able to repair harmful mutations or, when the damage is beyond repair, to trigger cell death. The immune system is also "trained" to recognize cancer cells and destroy them.
When all these defense mechanisms fail, cancer cells start dividing uncontrollably.
As a result, all cancer cells carry a number of somatic mutations that set them apart from healthy cells, and some tend to be the same across different cancer patients: for example, specific mutational patterns found in lung cancer have been attributed to tobacco exposure and were indeed reproduced in animal models. Another set of mutations has been attributed to UV exposure and has been found in skin cancers [1, 2].
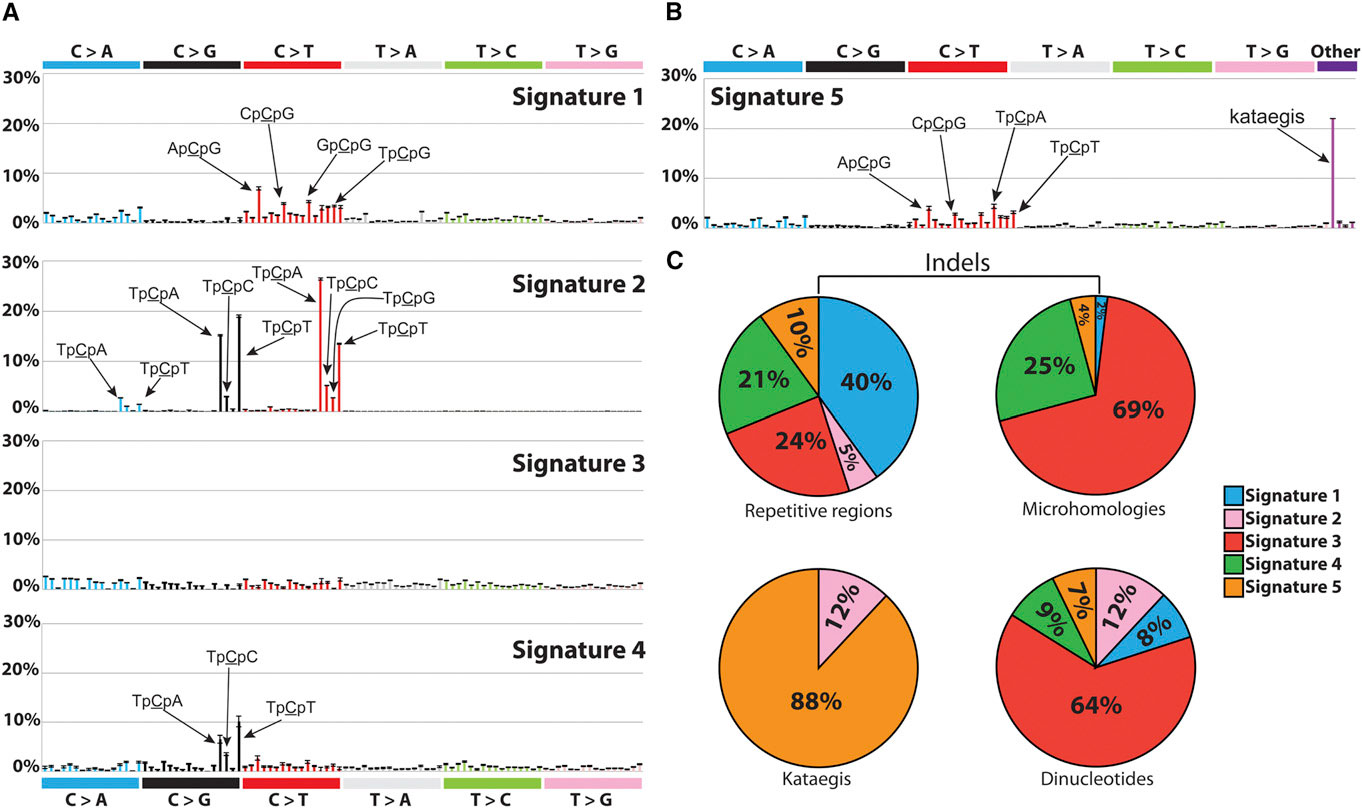
This prompts the ambitious question: can we find common mutations across individuals with the same cancer? And how many of these mutational patterns that are common across individuals can we attribute to particular exposures and/or biological processes? Distinguished postdoctoral researcher Ludmil Alexandrov, from the Los Alamos National Laboratory, has been working on this problem since his he was a PhD student at the Wellcome Trust Sanger Institute.
"It's like lifting fingerprints," Alexandrov explains. "The mutations are the fingerprints, but now we have to do the investigative work and find the 'perpetrator', i.e., the carcinogens that caused them." During his graduate studies, under the supervision of Mike Stratton of the Wellcome Trust Sanger Institute, Alexandrov developed a mathematical model that, given the cancer genomes from a number of patients, is able to pick the "common signals" across the patients -- i.e. mutation patterns that are common across the patients -- and classify them into "signatures."
"When formulated mathematically," Alexandrov explains, "the question can be expressed as the classic 'cocktail party' problem, where multiple people in a room are speaking simultaneously while several microphones placed at different locations are recording the conversations. Each microphone captures a combination of all sounds and the problem is to identify the individual conversations from all the recordings." Taking from this analogy, each cancer genome is a "recording", and the task of the mathematical model is to reconstruct each conversation, in other words, the mutational patterns. These are sets of somatic mutations that are the observed across the cancer genomes and that characterize certain types of cancers.
In 2013, Alexandrov and colleagues analyzed 4,938,362 mutations from 7,042 patients, spanning 30 different cancers, and extracted more than 20 distinct mutational signatures [2]. "Some patterns were expected, like the known ones caused by tobacco and UV light," Alexandrov says. "Others were completely new."
Of the new signatures found, many are involved in defective DNA repair mechanisms, suggesting that drugs targeting these specific mechanisms may benefit cancers exhibiting these signatures [3]. But the most exciting part of this research will be finding the 'perpetrator' or, as Alexandrov explains, the mutations triggered by carcinogens like tobacco, UV radiation, obesity, and so on. The challenge will be to experimentally associate these mutational patterns to the exposures that caused them. In order to do this, the scientists will have to expose cultured cells and model organisms to known carcinogens and then analyze the genomes of the experimentally induced cancers.
In the meantime, the signatures found so far are only the beginning: Alexandrov and colleagues have teamed up with the Los Alamos High Performance Computing Organization in order to analyze the genomes of almost 30,000 cancer patients.
"The amount of data we will have to handle for this task is enormous, on the order of petabytes," Alexandrov says. "Few places in the world have the capability to handle this many data. Under normal circumstances, it takes months to answer a question on 10 petabytes of data. The supercomputing facility at Los Alamos can provide an answer within a day."
Because of his research, in 2014 Alexandrov was listed by Forbes magazine as one of the “30 brightest stars under the age of 30” in the field of Science and Healthcare. In 2015 he was awarded the AAAS Science & SciLifeLab Prize for Young Scientists in the category Genomics and Proteomics [2] and the Weintraub Award for Graduate Research. He is now the recipient of the prestigious Oppenheimer fellowship at Los Alamos National Laboratory.
References
Siegel, R., Miller, K., & Jemal, A. (2015). Cancer statistics, 2015 CA: A Cancer Journal for Clinicians, 65 (1), 5-29 DOI: 10.3322/caac.21254
[1] Alexandrov LB (2015). Understanding the origins of human cancer. Science (New York, N.Y.), 350 (6265) PMID: 26785464
[2] Alexandrov LB, Nik-Zainal S, Wedge DC, Aparicio SA, Behjati S, Biankin AV, Bignell GR, Bolli N, Borg A, Børresen-Dale AL, Boyault S, Burkhardt B, Butler AP, Caldas C, Davies HR, Desmedt C, Eils R, Eyfjörd JE, Foekens JA, Greaves M, Hosoda F, Hutter B, Ilicic T, Imbeaud S, Imielinski M, Jäger N, Jones DT, Jones D, Knappskog S, Kool M, Lakhani SR, López-Otín C, Martin S, Munshi NC, Nakamura H, Northcott PA, Pajic M, Papaemmanuil E, Paradiso A, Pearson JV, Puente XS, Raine K, Ramakrishna M, Richardson AL, Richter J, Rosenstiel P, Schlesner M, Schumacher TN, Span PN, Teague JW, Totoki Y, Tutt AN, Valdés-Mas R, van Buuren MM, van 't Veer L, Vincent-Salomon A, Waddell N, Yates LR, Australian Pancreatic Cancer Genome Initiative, ICGC Breast Cancer Consortium, ICGC MMML-Seq Consortium, ICGC PedBrain, Zucman-Rossi J, Futreal PA, McDermott U, Lichter P, Meyerson M, Grimmond SM, Siebert R, Campo E, Shibata T, Pfister SM, Campbell PJ, & Stratton MR (2013). Signatures of mutational processes in human cancer. Nature, 500 (7463), 415-21 PMID: 23945592
[3] Alexandrov LB, Nik-Zainal S, Siu HC, Leung SY, & Stratton MR (2015). A mutational signature in gastric cancer suggests therapeutic strategies. Nature communications, 6 PMID: 26511885
Labels:
cancer,
DNA,
genes,
genetic diseases,
genetic effects
Friday, April 22, 2016
Digging For Clues About Climate Change
Guest post by Rebecca McDonald, science writer
While many scientists who study climate change look up to the sky for clues about the Earth’s future, one researcher has spent her career looking down—at the abundance of life in the soil below. Innumerable microorganisms such as bacteria and fungi live in harmony with plant roots, decomposing fallen leaves and dead animals. In addition to acting as the ultimate recyclers, they also stabilize the soil and help to retain water.
Cheryl Kuske, a microbiologist at Los Alamos National Laboratory, has focused the last two and a half decades on studying this microbial environment. “By decomposing organic matter,” she explains, “microorganisms help cycle carbon and nitrogen through the ecosystem.” Some of the carbon and nitrogen released from the organic matter goes into the soil and is assimilated into roots to help new plants grow—the carbon is incorporated into sugars, and the nitrogen atoms are used to build proteins. But some of these molecules are also released as CO2 and N2 gases into the atmosphere.
The soil ecosystem functions in a delicate balance. Although some organisms release gases into the air, others—including certain bacteria and leafy plants—remove harmful CO2 from the atmosphere for food production.
Kuske and her colleagues at Los Alamos National Laboratory have been investigating the roles of these microbes in carbon and nitrogen cycling to help make better predictions about terrestrial ecosystem responses to climate change. Using a technique called metagenomics to sequence the DNA of all the microbes at once, the team can study the organisms’ genes and the enzymes they produce.
These microrganisms’ lifecycles are so intertwined that their single genomes cannot be isolated for sequencing. However, analyzed jointly, they yield important clues about their collective functions in the environment. Scientists can identify things such as which bacteria or fungi are responsible for fixing nitrogen or carbon, the ratio of bacteria to fungi in the soil, and which microbes are closely associated with root health or plant growth. The researchers can even figure out which enzymes are currently being used through a technique called meta-transcriptomics; this approach sequences only the transcripts of genomic data that are actively being made and used for protein synthesis.
By sampling microbes from various soil environments over long periods of time, Kuske’s team and collaborators are able to understand what happens under the surface when things change aboveground. For instance, in a recent long-term study in Utah, the scientists discovered that slight changes in the summer precipitation pattern, combined with a 2°C rise in soil temperature, resulted in significant changes in the population of microbes below: the types of organisms completely changed, thus altering their overall role in the environment. For example, cyanobacteria—bacteria that create energy through photosynthesis—were no longer present. As a consequence, the new population of microbes no longer had the ability to pull carbon out of the air and had a decreased capacity for fixing nitrogen for protein synthesis.
Increased nitrogen from industrial runoff or fertilizer from agriculture can also have significant effects on the composition of organisms in the soil, as nitrogen is an essential molecule for the growth of both plants and bacteria. A comparison of 15 recent field experiments where nitrogen deposition was measured showed that in an arid environment, an increase in nitrogen had a positive effect on soil health at low concentrations, but too much was toxic to the soil community [1]. In a field experiment in Nevada, higher nitrogen concentrations changed the species composition of bacteria—but not fungi—leading to a fungi-dominated community [2,3].
Although the ramifications of these changes to the microbial world are not yet completely understood, Kuske’s team is continuing their studies, both in the laboratory, under controlled conditions, as well as at various field sites in the American Southwest. What they do know is that the feedback loop is strong. Changes in the aboveground environment—such as rising temperatures, altered precipitation, and increased nitrogen runoff—lead to changes below ground that can have far-reaching consequences.
“The studies being conducted at Los Alamos provide an understanding of the interactive biological processes that are inherent in all types of terrestrial ecosystems and that tightly control carbon and nitrogen fluxes to the atmosphere,” says Kuske. Climate warming and altered weather patterns will disrupt this balance. When the diversity of soil microbes change, the feedback loops that ensue could have lasting effects on the amounts of carbon and nitrogen in the soil and the atmosphere.
Rebecca McDonald is a science writer at Los Alamos National Laboratory specializing in the communication of bioscience research. She has also worked as a freelance writer, and volunteers her time as a communications consultant for a science education non-profit.
Disclaimer: Elena E. Giorgi is a computational biologist in the Theoretical Division of the Los Alamos National Laboratory. She does not represent her employer’s views. LA-UR-16-22406.
[1] Steven B, Kuske CR, Gallegos-Graves LV, Reed SC, & Belnap J (2015). Climate change and physical disturbance manipulations result in distinct biological soil crust communities. Applied and environmental microbiology, 81 (21), 7448-59 PMID: 26276111
[2] Sinsabaugh RL, Belnap J, Rudgers J, Kuske CR, Martinez N, & Sandquist D (2015). Soil microbial responses to nitrogen addition in arid ecosystems. Frontiers in microbiology, 6 PMID: 26322030
[3] Mueller RC, Belnap J, & Kuske CR (2015). Soil bacterial and fungal community responses to nitrogen addition across soil depth and microhabitat in an arid shrubland. Frontiers in microbiology, 6 PMID: 26388845

 |
| Photo Credit: LeRoy N. Sanchez |
While many scientists who study climate change look up to the sky for clues about the Earth’s future, one researcher has spent her career looking down—at the abundance of life in the soil below. Innumerable microorganisms such as bacteria and fungi live in harmony with plant roots, decomposing fallen leaves and dead animals. In addition to acting as the ultimate recyclers, they also stabilize the soil and help to retain water.
Cheryl Kuske, a microbiologist at Los Alamos National Laboratory, has focused the last two and a half decades on studying this microbial environment. “By decomposing organic matter,” she explains, “microorganisms help cycle carbon and nitrogen through the ecosystem.” Some of the carbon and nitrogen released from the organic matter goes into the soil and is assimilated into roots to help new plants grow—the carbon is incorporated into sugars, and the nitrogen atoms are used to build proteins. But some of these molecules are also released as CO2 and N2 gases into the atmosphere.
The soil ecosystem functions in a delicate balance. Although some organisms release gases into the air, others—including certain bacteria and leafy plants—remove harmful CO2 from the atmosphere for food production.
Kuske and her colleagues at Los Alamos National Laboratory have been investigating the roles of these microbes in carbon and nitrogen cycling to help make better predictions about terrestrial ecosystem responses to climate change. Using a technique called metagenomics to sequence the DNA of all the microbes at once, the team can study the organisms’ genes and the enzymes they produce.
These microrganisms’ lifecycles are so intertwined that their single genomes cannot be isolated for sequencing. However, analyzed jointly, they yield important clues about their collective functions in the environment. Scientists can identify things such as which bacteria or fungi are responsible for fixing nitrogen or carbon, the ratio of bacteria to fungi in the soil, and which microbes are closely associated with root health or plant growth. The researchers can even figure out which enzymes are currently being used through a technique called meta-transcriptomics; this approach sequences only the transcripts of genomic data that are actively being made and used for protein synthesis.
 |
| Photo courtesy of Cheryl Kuske |
By sampling microbes from various soil environments over long periods of time, Kuske’s team and collaborators are able to understand what happens under the surface when things change aboveground. For instance, in a recent long-term study in Utah, the scientists discovered that slight changes in the summer precipitation pattern, combined with a 2°C rise in soil temperature, resulted in significant changes in the population of microbes below: the types of organisms completely changed, thus altering their overall role in the environment. For example, cyanobacteria—bacteria that create energy through photosynthesis—were no longer present. As a consequence, the new population of microbes no longer had the ability to pull carbon out of the air and had a decreased capacity for fixing nitrogen for protein synthesis.
Increased nitrogen from industrial runoff or fertilizer from agriculture can also have significant effects on the composition of organisms in the soil, as nitrogen is an essential molecule for the growth of both plants and bacteria. A comparison of 15 recent field experiments where nitrogen deposition was measured showed that in an arid environment, an increase in nitrogen had a positive effect on soil health at low concentrations, but too much was toxic to the soil community [1]. In a field experiment in Nevada, higher nitrogen concentrations changed the species composition of bacteria—but not fungi—leading to a fungi-dominated community [2,3].
Although the ramifications of these changes to the microbial world are not yet completely understood, Kuske’s team is continuing their studies, both in the laboratory, under controlled conditions, as well as at various field sites in the American Southwest. What they do know is that the feedback loop is strong. Changes in the aboveground environment—such as rising temperatures, altered precipitation, and increased nitrogen runoff—lead to changes below ground that can have far-reaching consequences.
“The studies being conducted at Los Alamos provide an understanding of the interactive biological processes that are inherent in all types of terrestrial ecosystems and that tightly control carbon and nitrogen fluxes to the atmosphere,” says Kuske. Climate warming and altered weather patterns will disrupt this balance. When the diversity of soil microbes change, the feedback loops that ensue could have lasting effects on the amounts of carbon and nitrogen in the soil and the atmosphere.
Rebecca McDonald is a science writer at Los Alamos National Laboratory specializing in the communication of bioscience research. She has also worked as a freelance writer, and volunteers her time as a communications consultant for a science education non-profit.
Disclaimer: Elena E. Giorgi is a computational biologist in the Theoretical Division of the Los Alamos National Laboratory. She does not represent her employer’s views. LA-UR-16-22406.
[1] Steven B, Kuske CR, Gallegos-Graves LV, Reed SC, & Belnap J (2015). Climate change and physical disturbance manipulations result in distinct biological soil crust communities. Applied and environmental microbiology, 81 (21), 7448-59 PMID: 26276111
[2] Sinsabaugh RL, Belnap J, Rudgers J, Kuske CR, Martinez N, & Sandquist D (2015). Soil microbial responses to nitrogen addition in arid ecosystems. Frontiers in microbiology, 6 PMID: 26322030
[3] Mueller RC, Belnap J, & Kuske CR (2015). Soil bacterial and fungal community responses to nitrogen addition across soil depth and microhabitat in an arid shrubland. Frontiers in microbiology, 6 PMID: 26388845
Labels:
bacteria,
climate change
Friday, April 8, 2016
The Antibacterial Resistance Threat: Are We Heading Toward a Post-Antibiotic Era?
 |
| Source: PEW Charitable Trusts |
The above graphic, from the Antibiotic Resistance Project by the PEW charitable trusts, summarizes how alarming the emergence of drug resistant bacterial strains has gotten over the past few decades. According to data from the Center for Disease Control (CDC), every year 2 million Americans acquire drug-resistant infections [1], in other words infections that do not respond to treatment with ordinary antibiotics. Not only do drug-resistant infections require much stronger drugs, but, when not deadly, they often leave patients with long-lasting complications.
One of the scariest threats is carbapenem-resistant Enterobacteriaceae (CRE), bacteria that are resistant to several kinds of antibiotics. In 2001, only North Carolina, out of all 50 states had reported one CRE infection. Last year, in 2015, 48 states reported CRE infections to the CDC. And while drug-resistant strains emerge rapidly, the discovery of antimicrobial substances has stalled: in the last decade, only 9 new antibiotics were approved, compared to 29 discovered in the 1980s and 23 in the 1990s. We are fighting a new war, and we are running out of weapons.
How does drug resistance emerge?
Bacteria constitute an irreplaceable building block of our ecosystem: they are found in soil, water, air, and in every living organism. In humans, it's estimated that they outnumber our cells by 3:1, and numerous studies have shown that not only do they help us digest and produce enzymes that our body wouldn't otherwise be able to break down, but they can also influence gene expression and certain phenotypes (see some of my past posts for more information).
They live in symbiosis with us, yet some bacteria can be highly pathogenic. The overall mortality rate from infectious diseases in the US fell by 75% over the first 15 years following the discovery of antibiotics [3], and researchers estimate that antibiotics have increased our lifespan by 2 to 10 years [4] by enabling us to fight infections that would otherwise be deadly.
However, evolution has taught bacteria to fight back.
Bacteria develop drug resistance through the acquisition of genetic mutations that either modify the bacteria's binding sites (and therefore the drug can no longer enter the membrane), or reduce the accumulation of the drug inside the bacterium. The latter happens through proteins called "efflux pumps", so called because their function is to pump drugs and other potentially harmful chemicals out of the cell. Once these advantageous mutations appear in the population, they spread very quickly, not only because they are selected for, but also thanks to bacteria's ability to transfer genes: the drug-resistant genes form a circular DNA unit called plasmid, and the unit is passed on to nearby bacteria so that they, too, can become drug resistant.
These mechanisms are not new to bacteria, however, what's new is the increasing overuse of antibiotics and antimicrobial chemicals in our modern lifestyle. The antimicrobial agent called triclosan, for example, can be found in all antibacterial soaps, toothpaste, mouthwash, detergents, and even toys and kitchen utensils. Because of its wide use in household and hygiene products, triclosan has been found in water, both natural streams and treated wastewater, as well as human samples of blood, urine, and breast milk. As though that alone wasn't enough to alert consumers, a study published on the Proceedings of the National Academy of Sciences [5] claims that triclosan, which can be absorbed through the skin, can impair the functioning of both skeletal and cardiac muscle. The researchers confirmed these findings both in vitro and in animal models.
Resistance is also spread through the use of antibiotics in industrial farming. In the US alone, the daily consumption of antibiotics amounts to 51 tons, of which around 80% is used in livestock, a little under 20% is for human use, and the rest is split between crops, pets, and aquaculture [3]. A meta-analysis published last year in PNAS [6] found that between 2000 and 2010 the global use of antibiotic drugs increased by 36%, with 76% of the increase coming from developing countries. The researchers projected that worldwide antibiotic consumption would rise by 67% by 2030 due to population growth and the increase in consumer demand.
These frightening statistics prompted CDC director Tom Frieden to issue a warning: “If we are not careful, we will soon be in a post-antibiotic era.” An era when common infections are deadly again.
"We need to be very careful in using antimicrobial agents for everything from hand washing to toothpaste," Harshini Mukundan, microbiologist at Los Alamos National Laboratory, explains. "Increased selection of drug resistant organisms means that future generations will be helpless in fighting even the most common bacterial infections."
Mukundan and her colleagues have been working on biosurveillance and tracking the emergence of drug resistant strains in high disease burden populations where emerging antibiotic resistance is a huge concern. In collaboration with the Los Alamos National Laboratory metagenomics group, and Los Alamos scientists Ben McMahon and Norman Doggett, the team is working on developing new assays for faster diagnosis of drug resistant infections. Another approach to fight drug resistance is trying to understand how bacterial efflux pumps work at excreting the drug out of the bacterium. Gnana Gnanakaran, a computational biologist at Los Alamos National Laboratory, and his team have developed mathematical models to describe the structure of these pumps [7] and find a way to deactivate them.
While this research is highly promising and exciting, we all need to step up and do our part before it's too late: the CDC published a series of recommendations for patients to follow at the doctor's office, and there are smart choices we can make at home, too. In a recent report, the Food and Drug Administration (FDA) claims that there is no evidence that antibacterial soaps do a better job at preventing infections than ordinary soap, and that in fact:
"New data suggest that the risks associated with long-term, daily use of antibacterial soaps may outweigh the benefits."In its 2011 policy paper, the Infectious Diseases Society of America (IDSA) recommended a substantial reduction in the use of antibiotics for growth promotion and feed efficiency in animal agriculture, and encouraged the FDA to complete and publish risk assessments of antibiotics currently approved for non-therapeutic use.
Just like any other precious resource, antibiotics (and antimicrobial drugs in general) need to be used with parsimony.
Resources:
[1] Antibiotic Resistance Threats in the United States, 2013 (CDC)
[2] PEW Antibiotic Resistance Poject
[3] Armstrong GL, Conn LA, & Pinner RW (1999). Trends in infectious disease mortality in the United States during the 20th century. JAMA, 281 (1), 61-6 PMID: 9892452
[4] Hollis, A., & Ahmed, Z. (2013). Preserving Antibiotics, Rationally New England Journal of Medicine, 369 (26), 2474-2476 DOI: 10.1056/NEJMp1311479
[5] Cherednichenko, G., Zhang, R., Bannister, R., Timofeyev, V., Li, N., Fritsch, E., Feng, W., Barrientos, G., Schebb, N., Hammock, B., Beam, K., Chiamvimonvat, N., & Pessah, I. (2012). Triclosan impairs excitation-contraction coupling and Ca2+ dynamics in striated muscle Proceedings of the National Academy of Sciences, 109 (35), 14158-14163 DOI: 10.1073/pnas.1211314109
[6] Van Boeckel, T., Brower, C., Gilbert, M., Grenfell, B., Levin, S., Robinson, T., Teillant, A., & Laxminarayan, R. (2015). Global trends in antimicrobial use in food animals Proceedings of the National Academy of Sciences, 112 (18), 5649-5654 DOI: 10.1073/pnas.1503141112
[7] Resisting Bacterial Resistance, by Rebecca McDonald, 1663 Magazine.
Labels:
allergies,
bacteria,
climate change,
drugs
Subscribe to:
Comments (Atom)















